Maxwell DeNies
PhD ’19, Cell and Molecular Biology/Mechanical Engineering

Cells are composed of highly interconnect complex networks of proteins, nucleic acids and metabolites. Within these networks, proteins are largely responsible for cellular function. To better understand which proteins are involved in specific pathways and how they modulate cell function, we use mass spectrometry based proteomics to identify proteins and quantify their abundance. However due to cellular complexity, traditional proteomic data can be difficult to interpret and is fundamentally limited by temporal resolution.
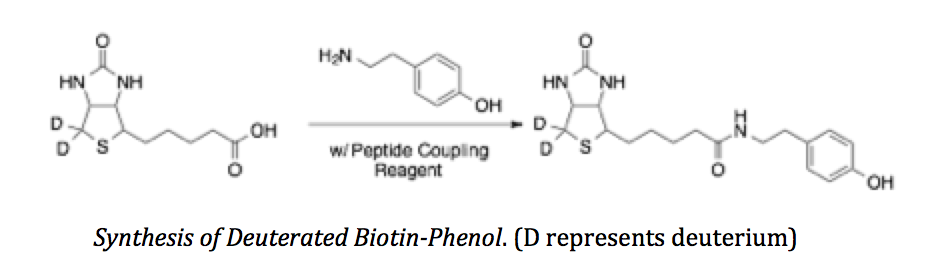
In this proposal, we will build upon recent technological advancements in spatially resolved proteomics and use our knowledge of chemical isotopes to design novel compounds to add temporal resolution to mass spectrometry based proteomics experiments. Similarly to live cell imaging, the addition of temporal resolution to our experiments will allow us to study how protein dynamics change overtime—without the visual constraints associated with microscopy.
In collaboration with the chemistry department, we will develop a synthesis strategy to create structurally identical chemical compounds with different masses that we can distinguish by mass spectrometry. Consequently, by enzymatically tagging proteins with differentially massed compounds we will add temporal resolution to mass spectrometry based proteomics. We believe that our new technology will be widely applicable in cell biology research.
Library Mentor: Ye Li